This page was generated from /home/docs/checkouts/readthedocs.org/user_builds/oceanspy/checkouts/latest/docs/Particles.ipynb.
Use Case 2: Particles
In this example we will subsample a dataset stored on SciServer using trajectories of synthetic Lagrangian particles. OceanSpy enables the extraction of properties at any given location, but it does not have functionality to compute Lagrangian trajectories yet. The trajectories used in this notebook have been previously computed using a Matlab-based Lagrangian particle code (Gelderloos et al., 2016). The Lagrangian particle code, along with synthetic oceanographic observational platforms (such as Argo floats, isobaric floats, current profiler, and gliders), will be incorporated into OceanSpy in future releases.
The animation below shows the change in temperature of synthetic particles along their 3D trajectories. Particles have been released in Denmark Strait and tracked backward in time from day 0 to day 180, and tracked forward in time from day 180 to 360. All particles are animated forward in time. Source: https://player.vimeo.com/video/296949375
[1]:
# Display animation
from IPython.display import HTML
# Link
s = "https://player.vimeo.com/video/296949375"
# Options
w = 640
h = 360
f = 0
a = "autoplay; fullscreen"
# Display
HTML(
f'<iframe src="{s}"'
'width="640" height="360" frameborder="0" allow="autoplay; fullscreen"'
"allowfullscreen></iframe>"
)
/home/idies/mambaforge/envs/Oceanography/lib/python3.9/site-packages/IPython/core/display.py:431: UserWarning: Consider using IPython.display.IFrame instead
warnings.warn("Consider using IPython.display.IFrame instead")
[1]:

Extract
The computational time to extract particle properties depends on the number of particles, the length of their trajectories, the number of variables requested, and, more importantly, how far the particles travel in the domain and therefore, how dispersed the Lagrangian trajectories are. Thus, it is preferable to extract particle properties asynchronously using the Job mode of SciServer compute (see SciServer access for step-by-step instructions). This
notebook, when executed in interactive mode, skips the subsampling step. Instead, it loads an OceanDataset previously created executing the same notebook in Job mode.
Set interactive = True to run in Interactive mode, or interactive = False to run in Job mode. Alternatively, you can activate the Job mode by typing interactive = False in the Parameters slot of the SciServer Job submission form.
[2]:
interactive = True # True: Interactive - False: Job
# Check parameters.txt
try:
# Use values from parameters.txt
f = open("parameters.txt", "r")
for line in f:
exec(line)
f.close()
except FileNotFoundError:
# Keep preset values
pass
The following cell starts a dask client (see the Dask Client section in the tutorial).
[3]:
# Start Client
from dask.distributed import Client
client = Client()
client
[3]:
Client
Client-ac9f71bc-d8db-11ed-8a48-0242ac110006
| Connection method: Cluster object | Cluster type: distributed.LocalCluster |
| Dashboard: http://127.0.0.1:8787/status |
Cluster Info
LocalCluster
fd40e064
| Dashboard: http://127.0.0.1:8787/status | Workers: 4 |
| Total threads: 4 | Total memory: 100.00 GiB |
| Status: running | Using processes: True |
Scheduler Info
Scheduler
Scheduler-ba5c5b4a-57ca-450f-a56f-6a9529bc98c5
| Comm: tcp://127.0.0.1:40685 | Workers: 4 |
| Dashboard: http://127.0.0.1:8787/status | Total threads: 4 |
| Started: Just now | Total memory: 100.00 GiB |
Workers
Worker: 0
| Comm: tcp://127.0.0.1:37562 | Total threads: 1 |
| Dashboard: http://127.0.0.1:39030/status | Memory: 25.00 GiB |
| Nanny: tcp://127.0.0.1:44469 | |
| Local directory: /tmp/dask-worker-space/worker-5eczcgfc | |
Worker: 1
| Comm: tcp://127.0.0.1:40873 | Total threads: 1 |
| Dashboard: http://127.0.0.1:38400/status | Memory: 25.00 GiB |
| Nanny: tcp://127.0.0.1:34121 | |
| Local directory: /tmp/dask-worker-space/worker-0p2ot6wv | |
Worker: 2
| Comm: tcp://127.0.0.1:33771 | Total threads: 1 |
| Dashboard: http://127.0.0.1:36647/status | Memory: 25.00 GiB |
| Nanny: tcp://127.0.0.1:33261 | |
| Local directory: /tmp/dask-worker-space/worker-og03lnsb | |
Worker: 3
| Comm: tcp://127.0.0.1:40242 | Total threads: 1 |
| Dashboard: http://127.0.0.1:41705/status | Memory: 25.00 GiB |
| Nanny: tcp://127.0.0.1:38136 | |
| Local directory: /tmp/dask-worker-space/worker-8yonp5i0 | |
While the following cell set up the environment.
[4]:
import matplotlib.pyplot as plt
import numpy as np
# Additional imports
import xarray as xr
from cartopy.crs import PlateCarree
# Import OceanSpy
import oceanspy as ospy
Here we open the OceanDataset containing the Eulerian fields that we want to sample along the Lagrangian trajectories. Then, for plotting purposes, we mask out the land from the bathymetry (variable Depth) and merge the variable masked_Depth into the OceanDataset.
[5]:
# Open OceanDataset (eulerian fields)
od_eul = ospy.open_oceandataset.from_catalog("EGshelfIIseas2km_ASR_full")
# Mask depth for plotting purposes
Depth = od_eul.dataset["Depth"]
Depth = Depth.where(Depth > 0)
od_eul = od_eul.merge_into_oceandataset(Depth.rename("masked_Depth"))
Opening EGshelfIIseas2km_ASR_full.
High-resolution (~2km) numerical simulation covering the east Greenland shelf (EGshelf),
and the Iceland and Irminger Seas (IIseas) forced by the Arctic System Reanalysis (ASR).
Citation:
* Almansi et al., 2020 - GRL.
Characteristics:
* full: Full domain without variables to close budgets.
See also:
* EGshelfIIseas2km_ASR_crop: Cropped domain with variables to close budgets.
We will analyze about 4,000 synthetic particles released in Denmark Strait at the end of February, and tracked forward in time for 30 days (with 6 hour resolution). The following properties will be extracted: temperature, salinity, ocean depth, and the vertical component of relative vorticity. Note that OceanSpy can extract any property, and variables such as momVort3 that are not in the model cell centers can be subsampled.
[6]:
import os
import subprocess
# Create or download OceanDataset with particle properties
mat_name = "oceanspy_particle_trajectories.mat" # Used by Job
nc_name = "oceanspy_particle_properties.nc" # Created by Job
if interactive:
# Download OceanDataset
if not os.path.isdir(mat_name):
import subprocess
print(f"Downloading [{nc_name}].")
commands = [
f"wget -v -O {nc_name} -L "
"https://livejohnshopkins-my.sharepoint.com/"
":u:/g/personal/malmans2_jh_edu/"
"EWvf_TyoEdpaDKcFacaPLI4B1fLGf9qleW7xbIDlKVPJDw?"
"download=1"
]
subprocess.call("&&".join(commands), shell=True)
else:
# Download trajectories
if not os.path.isdir(mat_name):
print(f"Downloading [{mat_name}].")
commands = [
f"wget -v -O {mat_name} -L "
"https://livejohnshopkins-my.sharepoint.com/"
":u:/g/personal/malmans2_jh_edu/"
"ETSRG8OcbWpccc7zx_rbzsIBqBl1UATQSwNTjqtk9fLR-Q?"
"download=1"
]
subprocess.call("&&".join(commands), shell=True)
# Read trajectories
import scipy.io as sio
particle_data = sio.loadmat(mat_name)
# Get time
time_origin = np.datetime64("2008-02-29T00:00")
times = np.asarray(
[
time_origin + np.timedelta64(int(dt * 24), "h")
for dt in particle_data["timee"].squeeze()
]
)
# Get trajectories
Xpart = particle_data["f_lons"].transpose()
Ypart = particle_data["f_lats"].transpose()
Zpart = particle_data["f_deps"].transpose()
# Pick variables (velocities and hydrography)
varList = ["Temp", "S", "momVort3", "Depth"]
# Extract properties
od_lag = ospy.subsample.particle_properties(
od_eul,
times=times,
Ypart=Ypart,
Xpart=Xpart,
Zpart=Zpart,
varList=varList,
)
# Save in netCDF format
od_lag.to_netcdf(nc_name)
# Open OceanDataset
od_lag = ospy.open_oceandataset.from_netcdf(nc_name)
Downloading [oceanspy_particle_properties.nc].
--2023-04-11 22:44:06-- https://livejohnshopkins-my.sharepoint.com/:u:/g/personal/malmans2_jh_edu/EWvf_TyoEdpaDKcFacaPLI4B1fLGf9qleW7xbIDlKVPJDw?download=1
Resolving livejohnshopkins-my.sharepoint.com (livejohnshopkins-my.sharepoint.com)... 13.107.138.8, 13.107.136.8, 2620:1ec:8f8::8, ...
Connecting to livejohnshopkins-my.sharepoint.com (livejohnshopkins-my.sharepoint.com)|13.107.138.8|:443... connected.
HTTP request sent, awaiting response... 302 Found
Location: /personal/malmans2_jh_edu/Documents/BoxMigration/oceanspy_particle_properties.nc?ga=1 [following]
--2023-04-11 22:44:07-- https://livejohnshopkins-my.sharepoint.com/personal/malmans2_jh_edu/Documents/BoxMigration/oceanspy_particle_properties.nc?ga=1
Reusing existing connection to livejohnshopkins-my.sharepoint.com:443.
HTTP request sent, awaiting response... 200 OK
Length: 45981653 (44M) [application/x-netcdf]
Saving to: ‘oceanspy_particle_properties.nc’
0K .......... .......... .......... .......... .......... 0% 166K 4m31s
50K .......... .......... .......... .......... .......... 0% 6.05M 2m19s
100K .......... .......... .......... .......... .......... 0% 11.6M 94s
150K .......... .......... .......... .......... .......... 0% 6.16M 72s
200K .......... .......... .......... .......... .......... 0% 5.79M 59s
250K .......... .......... .......... .......... .......... 0% 6.10M 50s
300K .......... .......... .......... .......... .......... 0% 11.4M 44s
350K .......... .......... .......... .......... .......... 0% 2.57M 40s
400K .......... .......... .......... .......... .......... 1% 6.13M 37s
450K .......... .......... .......... .......... .......... 1% 10.5M 33s
500K .......... .......... .......... .......... .......... 1% 5.65M 31s
550K .......... .......... .......... .......... .......... 1% 10.0M 29s
600K .......... .......... .......... .......... .......... 1% 5.55M 27s
650K .......... .......... .......... .......... .......... 1% 10.4M 25s
700K .......... .......... .......... .......... .......... 1% 5.71M 24s
750K .......... .......... .......... .......... .......... 1% 11.1M 23s
800K .......... .......... .......... .......... .......... 1% 10.5M 22s
850K .......... .......... .......... .......... .......... 2% 9.71M 21s
900K .......... .......... .......... .......... .......... 2% 6.04M 20s
950K .......... .......... .......... .......... .......... 2% 11.8M 19s
1000K .......... .......... .......... .......... .......... 2% 12.0M 18s
1050K .......... .......... .......... .......... .......... 2% 306K 24s
1100K .......... .......... .......... .......... .......... 2% 11.7M 23s
1150K .......... .......... .......... .......... .......... 2% 144K 35s
1200K .......... .......... .......... .......... .......... 2% 4.04M 34s
1250K .......... .......... .......... .......... .......... 2% 5.90M 33s
1300K .......... .......... .......... .......... .......... 3% 5.87M 32s
1350K .......... .......... .......... .......... .......... 3% 4.12M 31s
1400K .......... .......... .......... .......... .......... 3% 6.07M 30s
1450K .......... .......... .......... .......... .......... 3% 3.64M 29s
1500K .......... .......... .......... .......... .......... 3% 5.98M 29s
1550K .......... .......... .......... .......... .......... 3% 5.83M 28s
1600K .......... .......... .......... .......... .......... 3% 5.93M 27s
1650K .......... .......... .......... .......... .......... 3% 5.94M 27s
1700K .......... .......... .......... .......... .......... 3% 6.18M 26s
1750K .......... .......... .......... .......... .......... 4% 11.1M 25s
1800K .......... .......... .......... .......... .......... 4% 6.26M 25s
1850K .......... .......... .......... .......... .......... 4% 6.10M 24s
1900K .......... .......... .......... .......... .......... 4% 11.6M 24s
1950K .......... .......... .......... .......... .......... 4% 6.08M 23s
2000K .......... .......... .......... .......... .......... 4% 11.5M 23s
2050K .......... .......... .......... .......... .......... 4% 11.6M 22s
2100K .......... .......... .......... .......... .......... 4% 5.73M 22s
2150K .......... .......... .......... .......... .......... 4% 11.1M 22s
2200K .......... .......... .......... .......... .......... 5% 12.1M 21s
2250K .......... .......... .......... .......... .......... 5% 5.99M 21s
2300K .......... .......... .......... .......... .......... 5% 11.2M 20s
2350K .......... .......... .......... .......... .......... 5% 11.7M 20s
2400K .......... .......... .......... .......... .......... 5% 12.2M 20s
2450K .......... .......... .......... .......... .......... 5% 11.2M 19s
2500K .......... .......... .......... .......... .......... 5% 10.7M 19s
2550K .......... .......... .......... .......... .......... 5% 11.3M 19s
2600K .......... .......... .......... .......... .......... 5% 12.0M 18s
2650K .......... .......... .......... .......... .......... 6% 11.3M 18s
2700K .......... .......... .......... .......... .......... 6% 12.1M 18s
2750K .......... .......... .......... .......... .......... 6% 11.9M 18s
2800K .......... .......... .......... .......... .......... 6% 11.7M 17s
2850K .......... .......... .......... .......... .......... 6% 11.8M 17s
2900K .......... .......... .......... .......... .......... 6% 12.0M 17s
2950K .......... .......... .......... .......... .......... 6% 12.2M 17s
3000K .......... .......... .......... .......... .......... 6% 11.7M 16s
3050K .......... .......... .......... .......... .......... 6% 11.3M 16s
3100K .......... .......... .......... .......... .......... 7% 12.7M 16s
3150K .......... .......... .......... .......... .......... 7% 12.1M 16s
3200K .......... .......... .......... .......... .......... 7% 12.2M 15s
3250K .......... .......... .......... .......... .......... 7% 13.0M 15s
3300K .......... .......... .......... .......... .......... 7% 10.9M 15s
3350K .......... .......... .......... .......... .......... 7% 49.5M 15s
3400K .......... .......... .......... .......... .......... 7% 255K 17s
3450K .......... .......... .......... .......... .......... 7% 29.7K 37s
3500K .......... .......... .......... .......... .......... 7% 45.3K 49s
3550K .......... .......... .......... .......... .......... 8% 74.0K 56s
3600K .......... .......... .......... .......... .......... 8% 1.50M 55s
3650K .......... .......... .......... .......... .......... 8% 2.35M 55s
3700K .......... .......... .......... .......... .......... 8% 3.82M 54s
3750K .......... .......... .......... .......... .......... 8% 5.64M 53s
3800K .......... .......... .......... .......... .......... 8% 5.78M 53s
3850K .......... .......... .......... .......... .......... 8% 5.67M 52s
3900K .......... .......... .......... .......... .......... 8% 11.1M 52s
3950K .......... .......... .......... .......... .......... 8% 10.8M 51s
4000K .......... .......... .......... .......... .......... 9% 11.7M 50s
4050K .......... .......... .......... .......... .......... 9% 11.1M 50s
4100K .......... .......... .......... .......... .......... 9% 11.7M 49s
4150K .......... .......... .......... .......... .......... 9% 12.0M 48s
4200K .......... .......... .......... .......... .......... 9% 11.5M 48s
4250K .......... .......... .......... .......... .......... 9% 10.8M 47s
4300K .......... .......... .......... .......... .......... 9% 11.7M 47s
4350K .......... .......... .......... .......... .......... 9% 11.8M 46s
4400K .......... .......... .......... .......... .......... 9% 11.6M 46s
4450K .......... .......... .......... .......... .......... 10% 12.2M 45s
4500K .......... .......... .......... .......... .......... 10% 11.9M 45s
4550K .......... .......... .......... .......... .......... 10% 12.4M 44s
4600K .......... .......... .......... .......... .......... 10% 11.4M 44s
4650K .......... .......... .......... .......... .......... 10% 12.2M 43s
4700K .......... .......... .......... .......... .......... 10% 12.0M 43s
4750K .......... .......... .......... .......... .......... 10% 57.2M 42s
4800K .......... .......... .......... .......... .......... 10% 12.4M 42s
4850K .......... .......... .......... .......... .......... 10% 11.7M 41s
4900K .......... .......... .......... .......... .......... 11% 11.7M 41s
4950K .......... .......... .......... .......... .......... 11% 12.5M 40s
5000K .......... .......... .......... .......... .......... 11% 66.5M 40s
5050K .......... .......... .......... .......... .......... 11% 12.2M 39s
5100K .......... .......... .......... .......... .......... 11% 12.6M 39s
5150K .......... .......... .......... .......... .......... 11% 12.6M 39s
5200K .......... .......... .......... .......... .......... 11% 12.2M 38s
5250K .......... .......... .......... .......... .......... 11% 59.9M 38s
5300K .......... .......... .......... .......... .......... 11% 13.1M 38s
5350K .......... .......... .......... .......... .......... 12% 304K 38s
5400K .......... .......... .......... .......... .......... 12% 6.14M 38s
5450K .......... .......... .......... .......... .......... 12% 38.5K 47s
5500K .......... .......... .......... .......... .......... 12% 123K 49s
5550K .......... .......... .......... .......... .......... 12% 107K 52s
5600K .......... .......... .......... .......... .......... 12% 42.5K 60s
5650K .......... .......... .......... .......... .......... 12% 31.4K 70s
5700K .......... .......... .......... .......... .......... 12% 195K 71s
5750K .......... .......... .......... .......... .......... 12% 1.36M 71s
5800K .......... .......... .......... .......... .......... 13% 3.86M 70s
5850K .......... .......... .......... .......... .......... 13% 2.88M 69s
5900K .......... .......... .......... .......... .......... 13% 10.9M 69s
5950K .......... .......... .......... .......... .......... 13% 5.83M 68s
6000K .......... .......... .......... .......... .......... 13% 11.4M 68s
6050K .......... .......... .......... .......... .......... 13% 11.2M 67s
6100K .......... .......... .......... .......... .......... 13% 11.4M 66s
6150K .......... .......... .......... .......... .......... 13% 12.0M 66s
6200K .......... .......... .......... .......... .......... 13% 77.7M 65s
6250K .......... .......... .......... .......... .......... 14% 11.6M 65s
6300K .......... .......... .......... .......... .......... 14% 11.8M 64s
6350K .......... .......... .......... .......... .......... 14% 12.7M 63s
6400K .......... .......... .......... .......... .......... 14% 11.9M 63s
6450K .......... .......... .......... .......... .......... 14% 65.4M 62s
6500K .......... .......... .......... .......... .......... 14% 12.1M 62s
6550K .......... .......... .......... .......... .......... 14% 13.5M 61s
6600K .......... .......... .......... .......... .......... 14% 4.41M 61s
6650K .......... .......... .......... .......... .......... 14% 12.5M 60s
6700K .......... .......... .......... .......... .......... 15% 80.1M 60s
6750K .......... .......... .......... .......... .......... 15% 12.3M 59s
6800K .......... .......... .......... .......... .......... 15% 14.3M 59s
6850K .......... .......... .......... .......... .......... 15% 21.1M 58s
6900K .......... .......... .......... .......... .......... 15% 21.2M 58s
6950K .......... .......... .......... .......... .......... 15% 13.6M 57s
7000K .......... .......... .......... .......... .......... 15% 4.47M 57s
7050K .......... .......... .......... .......... .......... 15% 12.2M 56s
7100K .......... .......... .......... .......... .......... 15% 82.3M 56s
7150K .......... .......... .......... .......... .......... 16% 13.3M 56s
7200K .......... .......... .......... .......... .......... 16% 13.3M 55s
7250K .......... .......... .......... .......... .......... 16% 37.8M 55s
7300K .......... .......... .......... .......... .......... 16% 16.4M 54s
7350K .......... .......... .......... .......... .......... 16% 62.5M 54s
7400K .......... .......... .......... .......... .......... 16% 14.3M 53s
7450K .......... .......... .......... .......... .......... 16% 14.3M 53s
7500K .......... .......... .......... .......... .......... 16% 46.9M 53s
7550K .......... .......... .......... .......... .......... 16% 3.70M 52s
7600K .......... .......... .......... .......... .......... 17% 72.6M 52s
7650K .......... .......... .......... .......... .......... 17% 13.2M 51s
7700K .......... .......... .......... .......... .......... 17% 51.7M 51s
7750K .......... .......... .......... .......... .......... 17% 13.2M 51s
7800K .......... .......... .......... .......... .......... 17% 78.5M 50s
7850K .......... .......... .......... .......... .......... 17% 14.0M 50s
7900K .......... .......... .......... .......... .......... 17% 14.6M 50s
7950K .......... .......... .......... .......... .......... 17% 62.0M 49s
8000K .......... .......... .......... .......... .......... 17% 14.1M 49s
8050K .......... .......... .......... .......... .......... 18% 70.6M 48s
8100K .......... .......... .......... .......... .......... 18% 13.4M 48s
8150K .......... .......... .......... .......... .......... 18% 14.6M 48s
8200K .......... .......... .......... .......... .......... 18% 70.3M 47s
8250K .......... .......... .......... .......... .......... 18% 13.2M 47s
8300K .......... .......... .......... .......... .......... 18% 75.8M 47s
8350K .......... .......... .......... .......... .......... 18% 12.8M 46s
8400K .......... .......... .......... .......... .......... 18% 398K 47s
8450K .......... .......... .......... .......... .......... 18% 58.5M 46s
8500K .......... .......... .......... .......... .......... 19% 14.3M 46s
8550K .......... .......... .......... .......... .......... 19% 9.64M 46s
8600K .......... .......... .......... .......... .......... 19% 16.7M 45s
8650K .......... .......... .......... .......... .......... 19% 12.0M 45s
8700K .......... .......... .......... .......... .......... 19% 12.9M 45s
8750K .......... .......... .......... .......... .......... 19% 12.8M 44s
8800K .......... .......... .......... .......... .......... 19% 60.3M 44s
8850K .......... .......... .......... .......... .......... 19% 11.9M 44s
8900K .......... .......... .......... .......... .......... 19% 5.60M 44s
8950K .......... .......... .......... .......... .......... 20% 77.6M 43s
9000K .......... .......... .......... .......... .......... 20% 12.2M 43s
9050K .......... .......... .......... .......... .......... 20% 11.2M 43s
9100K .......... .......... .......... .......... .......... 20% 12.3M 42s
9150K .......... .......... .......... .......... .......... 20% 12.7M 42s
9200K .......... .......... .......... .......... .......... 20% 68.1M 42s
9250K .......... .......... .......... .......... .......... 20% 11.8M 42s
9300K .......... .......... .......... .......... .......... 20% 10.6M 41s
9350K .......... .......... .......... .......... .......... 20% 13.0M 41s
9400K .......... .......... .......... .......... .......... 21% 58.0M 41s
9450K .......... .......... .......... .......... .......... 21% 11.7M 41s
9500K .......... .......... .......... .......... .......... 21% 12.6M 40s
9550K .......... .......... .......... .......... .......... 21% 9.80M 40s
9600K .......... .......... .......... .......... .......... 21% 19.0M 40s
9650K .......... .......... .......... .......... .......... 21% 24.0M 40s
9700K .......... .......... .......... .......... .......... 21% 12.1M 39s
9750K .......... .......... .......... .......... .......... 21% 12.0M 39s
9800K .......... .......... .......... .......... .......... 21% 18.4M 39s
9850K .......... .......... .......... .......... .......... 22% 11.8M 39s
9900K .......... .......... .......... .......... .......... 22% 29.6M 38s
9950K .......... .......... .......... .......... .......... 22% 12.3M 38s
10000K .......... .......... .......... .......... .......... 22% 14.8M 38s
10050K .......... .......... .......... .......... .......... 22% 14.2M 38s
10100K .......... .......... .......... .......... .......... 22% 23.2M 37s
10150K .......... .......... .......... .......... .......... 22% 14.4M 37s
10200K .......... .......... .......... .......... .......... 22% 16.1M 37s
10250K .......... .......... .......... .......... .......... 22% 2.32M 37s
10300K .......... .......... .......... .......... .......... 23% 11.0M 37s
10350K .......... .......... .......... .......... .......... 23% 12.1M 36s
10400K .......... .......... .......... .......... .......... 23% 12.5M 36s
10450K .......... .......... .......... .......... .......... 23% 53.3M 36s
10500K .......... .......... .......... .......... .......... 23% 12.8M 36s
10550K .......... .......... .......... .......... .......... 23% 11.8M 36s
10600K .......... .......... .......... .......... .......... 23% 62.3M 35s
10650K .......... .......... .......... .......... .......... 23% 12.5M 35s
10700K .......... .......... .......... .......... .......... 23% 12.1M 35s
10750K .......... .......... .......... .......... .......... 24% 14.9M 35s
10800K .......... .......... .......... .......... .......... 24% 56.9M 34s
10850K .......... .......... .......... .......... .......... 24% 12.3M 34s
10900K .......... .......... .......... .......... .......... 24% 12.7M 34s
10950K .......... .......... .......... .......... .......... 24% 42.3M 34s
11000K .......... .......... .......... .......... .......... 24% 16.4M 34s
11050K .......... .......... .......... .......... .......... 24% 12.2M 34s
11100K .......... .......... .......... .......... .......... 24% 39.4M 33s
11150K .......... .......... .......... .......... .......... 24% 12.6M 33s
11200K .......... .......... .......... .......... .......... 25% 15.3M 33s
11250K .......... .......... .......... .......... .......... 25% 54.2M 33s
11300K .......... .......... .......... .......... .......... 25% 13.0M 33s
11350K .......... .......... .......... .......... .......... 25% 68.2M 32s
11400K .......... .......... .......... .......... .......... 25% 13.8M 32s
11450K .......... .......... .......... .......... .......... 25% 11.9M 32s
11500K .......... .......... .......... .......... .......... 25% 13.2M 32s
11550K .......... .......... .......... .......... .......... 25% 61.5M 32s
11600K .......... .......... .......... .......... .......... 25% 13.9M 31s
11650K .......... .......... .......... .......... .......... 26% 58.0M 31s
11700K .......... .......... .......... .......... .......... 26% 13.8M 31s
11750K .......... .......... .......... .......... .......... 26% 4.29M 31s
11800K .......... .......... .......... .......... .......... 26% 61.8M 31s
11850K .......... .......... .......... .......... .......... 26% 13.6M 31s
11900K .......... .......... .......... .......... .......... 26% 12.1M 30s
11950K .......... .......... .......... .......... .......... 26% 72.3M 30s
12000K .......... .......... .......... .......... .......... 26% 13.8M 30s
12050K .......... .......... .......... .......... .......... 26% 69.5M 30s
12100K .......... .......... .......... .......... .......... 27% 12.1M 30s
12150K .......... .......... .......... .......... .......... 27% 71.7M 30s
12200K .......... .......... .......... .......... .......... 27% 4.00M 30s
12250K .......... .......... .......... .......... .......... 27% 13.7M 29s
12300K .......... .......... .......... .......... .......... 27% 70.1M 29s
12350K .......... .......... .......... .......... .......... 27% 11.4M 29s
12400K .......... .......... .......... .......... .......... 27% 12.9M 29s
12450K .......... .......... .......... .......... .......... 27% 16.3M 29s
12500K .......... .......... .......... .......... .......... 27% 27.6M 29s
12550K .......... .......... .......... .......... .......... 28% 13.1M 28s
12600K .......... .......... .......... .......... .......... 28% 13.1M 28s
12650K .......... .......... .......... .......... .......... 28% 12.2M 28s
12700K .......... .......... .......... .......... .......... 28% 15.3M 28s
12750K .......... .......... .......... .......... .......... 28% 30.9M 28s
12800K .......... .......... .......... .......... .......... 28% 13.1M 28s
12850K .......... .......... .......... .......... .......... 28% 15.5M 28s
12900K .......... .......... .......... .......... .......... 28% 17.4M 27s
12950K .......... .......... .......... .......... .......... 28% 21.3M 27s
13000K .......... .......... .......... .......... .......... 29% 13.0M 27s
13050K .......... .......... .......... .......... .......... 29% 12.5M 27s
13100K .......... .......... .......... .......... .......... 29% 24.1M 27s
13150K .......... .......... .......... .......... .......... 29% 19.9M 27s
13200K .......... .......... .......... .......... .......... 29% 13.6M 27s
13250K .......... .......... .......... .......... .......... 29% 19.7M 26s
13300K .......... .......... .......... .......... .......... 29% 20.4M 26s
13350K .......... .......... .......... .......... .......... 29% 15.0M 26s
13400K .......... .......... .......... .......... .......... 29% 22.4M 26s
13450K .......... .......... .......... .......... .......... 30% 17.1M 26s
13500K .......... .......... .......... .......... .......... 30% 13.8M 26s
13550K .......... .......... .......... .......... .......... 30% 16.1M 26s
13600K .......... .......... .......... .......... .......... 30% 36.2M 26s
13650K .......... .......... .......... .......... .......... 30% 8.06M 25s
13700K .......... .......... .......... .......... .......... 30% 66.1M 25s
13750K .......... .......... .......... .......... .......... 30% 13.0M 25s
13800K .......... .......... .......... .......... .......... 30% 69.1M 25s
13850K .......... .......... .......... .......... .......... 30% 12.7M 25s
13900K .......... .......... .......... .......... .......... 31% 12.3M 25s
13950K .......... .......... .......... .......... .......... 31% 12.8M 25s
14000K .......... .......... .......... .......... .......... 31% 61.5M 25s
14050K .......... .......... .......... .......... .......... 31% 12.9M 24s
14100K .......... .......... .......... .......... .......... 31% 75.8M 24s
14150K .......... .......... .......... .......... .......... 31% 13.0M 24s
14200K .......... .......... .......... .......... .......... 31% 13.6M 24s
14250K .......... .......... .......... .......... .......... 31% 55.5M 24s
14300K .......... .......... .......... .......... .......... 31% 13.4M 24s
14350K .......... .......... .......... .......... .......... 32% 14.0M 24s
14400K .......... .......... .......... .......... .......... 32% 56.2M 24s
14450K .......... .......... .......... .......... .......... 32% 12.0M 23s
14500K .......... .......... .......... .......... .......... 32% 72.8M 23s
14550K .......... .......... .......... .......... .......... 32% 14.9M 23s
14600K .......... .......... .......... .......... .......... 32% 67.2M 23s
14650K .......... .......... .......... .......... .......... 32% 13.0M 23s
14700K .......... .......... .......... .......... .......... 32% 14.0M 23s
14750K .......... .......... .......... .......... .......... 32% 395K 23s
14800K .......... .......... .......... .......... .......... 33% 64.9M 23s
14850K .......... .......... .......... .......... .......... 33% 6.23M 23s
14900K .......... .......... .......... .......... .......... 33% 556K 23s
14950K .......... .......... .......... .......... .......... 33% 6.43M 23s
15000K .......... .......... .......... .......... .......... 33% 5.84M 23s
15050K .......... .......... .......... .......... .......... 33% 5.64M 23s
15100K .......... .......... .......... .......... .......... 33% 5.83M 23s
15150K .......... .......... .......... .......... .......... 33% 3.86M 22s
15200K .......... .......... .......... .......... .......... 33% 5.78M 22s
15250K .......... .......... .......... .......... .......... 34% 10.5M 22s
15300K .......... .......... .......... .......... .......... 34% 6.22M 22s
15350K .......... .......... .......... .......... .......... 34% 10.6M 22s
15400K .......... .......... .......... .......... .......... 34% 11.3M 22s
15450K .......... .......... .......... .......... .......... 34% 6.02M 22s
15500K .......... .......... .......... .......... .......... 34% 11.5M 22s
15550K .......... .......... .......... .......... .......... 34% 6.13M 22s
15600K .......... .......... .......... .......... .......... 34% 12.5M 22s
15650K .......... .......... .......... .......... .......... 34% 11.2M 21s
15700K .......... .......... .......... .......... .......... 35% 10.6M 21s
15750K .......... .......... .......... .......... .......... 35% 12.1M 21s
15800K .......... .......... .......... .......... .......... 35% 11.6M 21s
15850K .......... .......... .......... .......... .......... 35% 5.28M 21s
15900K .......... .......... .......... .......... .......... 35% 15.4M 21s
15950K .......... .......... .......... .......... .......... 35% 11.1M 21s
16000K .......... .......... .......... .......... .......... 35% 11.4M 21s
16050K .......... .......... .......... .......... .......... 35% 11.5M 21s
16100K .......... .......... .......... .......... .......... 35% 11.6M 21s
16150K .......... .......... .......... .......... .......... 36% 11.1M 21s
16200K .......... .......... .......... .......... .......... 36% 11.8M 20s
16250K .......... .......... .......... .......... .......... 36% 7.98M 20s
16300K .......... .......... .......... .......... .......... 36% 18.8M 20s
16350K .......... .......... .......... .......... .......... 36% 11.0M 20s
16400K .......... .......... .......... .......... .......... 36% 12.4M 20s
16450K .......... .......... .......... .......... .......... 36% 12.0M 20s
16500K .......... .......... .......... .......... .......... 36% 11.6M 20s
16550K .......... .......... .......... .......... .......... 36% 11.5M 20s
16600K .......... .......... .......... .......... .......... 37% 34.2M 20s
16650K .......... .......... .......... .......... .......... 37% 7.01M 20s
16700K .......... .......... .......... .......... .......... 37% 32.7M 20s
16750K .......... .......... .......... .......... .......... 37% 12.0M 19s
16800K .......... .......... .......... .......... .......... 37% 14.6M 19s
16850K .......... .......... .......... .......... .......... 37% 11.6M 19s
16900K .......... .......... .......... .......... .......... 37% 11.2M 19s
16950K .......... .......... .......... .......... .......... 37% 39.1M 19s
17000K .......... .......... .......... .......... .......... 37% 12.7M 19s
17050K .......... .......... .......... .......... .......... 38% 11.8M 19s
17100K .......... .......... .......... .......... .......... 38% 16.0M 19s
17150K .......... .......... .......... .......... .......... 38% 12.6M 19s
17200K .......... .......... .......... .......... .......... 38% 12.8M 19s
17250K .......... .......... .......... .......... .......... 38% 25.9M 19s
17300K .......... .......... .......... .......... .......... 38% 14.2M 19s
17350K .......... .......... .......... .......... .......... 38% 12.3M 18s
17400K .......... .......... .......... .......... .......... 38% 17.7M 18s
17450K .......... .......... .......... .......... .......... 38% 308K 19s
17500K .......... .......... .......... .......... .......... 39% 71.0M 18s
17550K .......... .......... .......... .......... .......... 39% 803K 18s
17600K .......... .......... .......... .......... .......... 39% 11.7M 18s
17650K .......... .......... .......... .......... .......... 39% 11.0M 18s
17700K .......... .......... .......... .......... .......... 39% 6.00M 18s
17750K .......... .......... .......... .......... .......... 39% 11.3M 18s
17800K .......... .......... .......... .......... .......... 39% 6.59M 18s
17850K .......... .......... .......... .......... .......... 39% 6.08M 18s
17900K .......... .......... .......... .......... .......... 39% 11.7M 18s
17950K .......... .......... .......... .......... .......... 40% 6.29M 18s
18000K .......... .......... .......... .......... .......... 40% 11.8M 18s
18050K .......... .......... .......... .......... .......... 40% 10.9M 18s
18100K .......... .......... .......... .......... .......... 40% 11.5M 18s
18150K .......... .......... .......... .......... .......... 40% 11.0M 18s
18200K .......... .......... .......... .......... .......... 40% 12.5M 17s
18250K .......... .......... .......... .......... .......... 40% 6.21M 17s
18300K .......... .......... .......... .......... .......... 40% 12.0M 17s
18350K .......... .......... .......... .......... .......... 40% 11.2M 17s
18400K .......... .......... .......... .......... .......... 41% 12.2M 17s
18450K .......... .......... .......... .......... .......... 41% 11.5M 17s
18500K .......... .......... .......... .......... .......... 41% 11.5M 17s
18550K .......... .......... .......... .......... .......... 41% 11.3M 17s
18600K .......... .......... .......... .......... .......... 41% 12.0M 17s
18650K .......... .......... .......... .......... .......... 41% 10.8M 17s
18700K .......... .......... .......... .......... .......... 41% 10.5M 17s
18750K .......... .......... .......... .......... .......... 41% 11.8M 17s
18800K .......... .......... .......... .......... .......... 41% 11.7M 17s
18850K .......... .......... .......... .......... .......... 42% 11.7M 17s
18900K .......... .......... .......... .......... .......... 42% 11.8M 16s
18950K .......... .......... .......... .......... .......... 42% 12.2M 16s
19000K .......... .......... .......... .......... .......... 42% 11.5M 16s
19050K .......... .......... .......... .......... .......... 42% 10.3M 16s
19100K .......... .......... .......... .......... .......... 42% 534K 16s
19150K .......... .......... .......... .......... .......... 42% 66.0M 16s
19200K .......... .......... .......... .......... .......... 42% 3.08M 16s
19250K .......... .......... .......... .......... .......... 42% 11.3M 16s
19300K .......... .......... .......... .......... .......... 43% 12.3M 16s
19350K .......... .......... .......... .......... .......... 43% 57.0M 16s
19400K .......... .......... .......... .......... .......... 43% 12.4M 16s
19450K .......... .......... .......... .......... .......... 43% 10.9M 16s
19500K .......... .......... .......... .......... .......... 43% 12.1M 16s
19550K .......... .......... .......... .......... .......... 43% 12.7M 16s
19600K .......... .......... .......... .......... .......... 43% 63.5M 16s
19650K .......... .......... .......... .......... .......... 43% 12.5M 16s
19700K .......... .......... .......... .......... .......... 43% 11.7M 16s
19750K .......... .......... .......... .......... .......... 44% 74.8M 15s
19800K .......... .......... .......... .......... .......... 44% 13.2M 15s
19850K .......... .......... .......... .......... .......... 44% 13.2M 15s
19900K .......... .......... .......... .......... .......... 44% 57.8M 15s
19950K .......... .......... .......... .......... .......... 44% 14.6M 15s
20000K .......... .......... .......... .......... .......... 44% 54.2M 15s
20050K .......... .......... .......... .......... .......... 44% 13.6M 15s
20100K .......... .......... .......... .......... .......... 44% 14.1M 15s
20150K .......... .......... .......... .......... .......... 44% 68.8M 15s
20200K .......... .......... .......... .......... .......... 45% 13.8M 15s
20250K .......... .......... .......... .......... .......... 45% 47.3M 15s
20300K .......... .......... .......... .......... .......... 45% 13.6M 15s
20350K .......... .......... .......... .......... .......... 45% 15.0M 15s
20400K .......... .......... .......... .......... .......... 45% 50.6M 15s
20450K .......... .......... .......... .......... .......... 45% 14.8M 15s
20500K .......... .......... .......... .......... .......... 45% 51.6M 15s
20550K .......... .......... .......... .......... .......... 45% 14.2M 14s
20600K .......... .......... .......... .......... .......... 45% 60.1M 14s
20650K .......... .......... .......... .......... .......... 46% 12.6M 14s
20700K .......... .......... .......... .......... .......... 46% 64.3M 14s
20750K .......... .......... .......... .......... .......... 46% 15.6M 14s
20800K .......... .......... .......... .......... .......... 46% 13.9M 14s
20850K .......... .......... .......... .......... .......... 46% 42.2M 14s
20900K .......... .......... .......... .......... .......... 46% 16.0M 14s
20950K .......... .......... .......... .......... .......... 46% 52.6M 14s
21000K .......... .......... .......... .......... .......... 46% 14.8M 14s
21050K .......... .......... .......... .......... .......... 46% 38.7M 14s
21100K .......... .......... .......... .......... .......... 47% 16.8M 14s
21150K .......... .......... .......... .......... .......... 47% 38.9M 14s
21200K .......... .......... .......... .......... .......... 47% 15.1M 14s
21250K .......... .......... .......... .......... .......... 47% 52.2M 14s
21300K .......... .......... .......... .......... .......... 47% 14.8M 14s
21350K .......... .......... .......... .......... .......... 47% 59.8M 13s
21400K .......... .......... .......... .......... .......... 47% 14.6M 13s
21450K .......... .......... .......... .......... .......... 47% 14.0M 13s
21500K .......... .......... .......... .......... .......... 47% 55.3M 13s
21550K .......... .......... .......... .......... .......... 48% 15.6M 13s
21600K .......... .......... .......... .......... .......... 48% 55.1M 13s
21650K .......... .......... .......... .......... .......... 48% 14.9M 13s
21700K .......... .......... .......... .......... .......... 48% 54.2M 13s
21750K .......... .......... .......... .......... .......... 48% 14.7M 13s
21800K .......... .......... .......... .......... .......... 48% 62.7M 13s
21850K .......... .......... .......... .......... .......... 48% 15.5M 13s
21900K .......... .......... .......... .......... .......... 48% 52.0M 13s
21950K .......... .......... .......... .......... .......... 48% 15.8M 13s
22000K .......... .......... .......... .......... .......... 49% 56.5M 13s
22050K .......... .......... .......... .......... .......... 49% 14.9M 13s
22100K .......... .......... .......... .......... .......... 49% 59.1M 13s
22150K .......... .......... .......... .......... .......... 49% 15.2M 13s
22200K .......... .......... .......... .......... .......... 49% 60.9M 13s
22250K .......... .......... .......... .......... .......... 49% 15.9M 12s
22300K .......... .......... .......... .......... .......... 49% 35.9M 12s
22350K .......... .......... .......... .......... .......... 49% 17.9M 12s
22400K .......... .......... .......... .......... .......... 49% 42.8M 12s
22450K .......... .......... .......... .......... .......... 50% 15.3M 12s
22500K .......... .......... .......... .......... .......... 50% 62.5M 12s
22550K .......... .......... .......... .......... .......... 50% 46.4M 12s
22600K .......... .......... .......... .......... .......... 50% 16.9M 12s
22650K .......... .......... .......... .......... .......... 50% 14.3M 12s
22700K .......... .......... .......... .......... .......... 50% 58.8M 12s
22750K .......... .......... .......... .......... .......... 50% 62.0M 12s
22800K .......... .......... .......... .......... .......... 50% 16.0M 12s
22850K .......... .......... .......... .......... .......... 50% 46.7M 12s
22900K .......... .......... .......... .......... .......... 51% 17.0M 12s
22950K .......... .......... .......... .......... .......... 51% 41.2M 12s
23000K .......... .......... .......... .......... .......... 51% 16.3M 12s
23050K .......... .......... .......... .......... .......... 51% 53.2M 12s
23100K .......... .......... .......... .......... .......... 51% 4.27M 12s
23150K .......... .......... .......... .......... .......... 51% 71.4M 12s
23200K .......... .......... .......... .......... .......... 51% 74.3M 12s
23250K .......... .......... .......... .......... .......... 51% 63.0M 11s
23300K .......... .......... .......... .......... .......... 51% 82.7M 11s
23350K .......... .......... .......... .......... .......... 52% 75.6M 11s
23400K .......... .......... .......... .......... .......... 52% 51.2M 11s
23450K .......... .......... .......... .......... .......... 52% 49.0M 11s
23500K .......... .......... .......... .......... .......... 52% 15.3M 11s
23550K .......... .......... .......... .......... .......... 52% 56.2M 11s
23600K .......... .......... .......... .......... .......... 52% 15.9M 11s
23650K .......... .......... .......... .......... .......... 52% 55.0M 11s
23700K .......... .......... .......... .......... .......... 52% 84.1M 11s
23750K .......... .......... .......... .......... .......... 53% 14.9M 11s
23800K .......... .......... .......... .......... .......... 53% 60.4M 11s
23850K .......... .......... .......... .......... .......... 53% 14.6M 11s
23900K .......... .......... .......... .......... .......... 53% 56.0M 11s
23950K .......... .......... .......... .......... .......... 53% 15.5M 11s
24000K .......... .......... .......... .......... .......... 53% 69.4M 11s
24050K .......... .......... .......... .......... .......... 53% 13.7M 11s
24100K .......... .......... .......... .......... .......... 53% 42.2M 11s
24150K .......... .......... .......... .......... .......... 53% 69.8M 11s
24200K .......... .......... .......... .......... .......... 54% 17.5M 11s
24250K .......... .......... .......... .......... .......... 54% 54.4M 11s
24300K .......... .......... .......... .......... .......... 54% 126K 11s
24350K .......... .......... .......... .......... .......... 54% 67.9M 11s
24400K .......... .......... .......... .......... .......... 54% 932K 11s
24450K .......... .......... .......... .......... .......... 54% 1.51M 11s
24500K .......... .......... .......... .......... .......... 54% 201K 11s
24550K .......... .......... .......... .......... .......... 54% 2.39M 11s
24600K .......... .......... .......... .......... .......... 54% 2.35M 11s
24650K .......... .......... .......... .......... .......... 55% 2.02M 11s
24700K .......... .......... .......... .......... .......... 55% 2.89M 11s
24750K .......... .......... .......... .......... .......... 55% 3.82M 11s
24800K .......... .......... .......... .......... .......... 55% 3.78M 11s
24850K .......... .......... .......... .......... .......... 55% 5.58M 11s
24900K .......... .......... .......... .......... .......... 55% 5.67M 11s
24950K .......... .......... .......... .......... .......... 55% 5.66M 11s
25000K .......... .......... .......... .......... .......... 55% 5.76M 11s
25050K .......... .......... .......... .......... .......... 55% 6.07M 10s
25100K .......... .......... .......... .......... .......... 56% 10.2M 10s
25150K .......... .......... .......... .......... .......... 56% 5.86M 10s
25200K .......... .......... .......... .......... .......... 56% 11.1M 10s
25250K .......... .......... .......... .......... .......... 56% 5.86M 10s
25300K .......... .......... .......... .......... .......... 56% 10.8M 10s
25350K .......... .......... .......... .......... .......... 56% 5.97M 10s
25400K .......... .......... .......... .......... .......... 56% 10.9M 10s
25450K .......... .......... .......... .......... .......... 56% 5.94M 10s
25500K .......... .......... .......... .......... .......... 56% 11.7M 10s
25550K .......... .......... .......... .......... .......... 57% 306K 10s
25600K .......... .......... .......... .......... .......... 57% 10.6M 10s
25650K .......... .......... .......... .......... .......... 57% 6.47M 10s
25700K .......... .......... .......... .......... .......... 57% 10.9M 10s
25750K .......... .......... .......... .......... .......... 57% 10.1M 10s
25800K .......... .......... .......... .......... .......... 57% 10.9M 10s
25850K .......... .......... .......... .......... .......... 57% 7.42M 10s
25900K .......... .......... .......... .......... .......... 57% 7.81M 10s
25950K .......... .......... .......... .......... .......... 57% 11.4M 10s
26000K .......... .......... .......... .......... .......... 58% 10.5M 10s
26050K .......... .......... .......... .......... .......... 58% 9.52M 10s
26100K .......... .......... .......... .......... .......... 58% 6.46M 10s
26150K .......... .......... .......... .......... .......... 58% 11.6M 10s
26200K .......... .......... .......... .......... .......... 58% 10.8M 10s
26250K .......... .......... .......... .......... .......... 58% 6.24M 10s
26300K .......... .......... .......... .......... .......... 58% 11.8M 10s
26350K .......... .......... .......... .......... .......... 58% 10.5M 10s
26400K .......... .......... .......... .......... .......... 58% 11.0M 9s
26450K .......... .......... .......... .......... .......... 59% 10.8M 9s
26500K .......... .......... .......... .......... .......... 59% 5.79M 9s
26550K .......... .......... .......... .......... .......... 59% 11.3M 9s
26600K .......... .......... .......... .......... .......... 59% 11.0M 9s
26650K .......... .......... .......... .......... .......... 59% 5.95M 9s
26700K .......... .......... .......... .......... .......... 59% 11.1M 9s
26750K .......... .......... .......... .......... .......... 59% 11.3M 9s
26800K .......... .......... .......... .......... .......... 59% 11.3M 9s
26850K .......... .......... .......... .......... .......... 59% 10.9M 9s
26900K .......... .......... .......... .......... .......... 60% 6.35M 9s
26950K .......... .......... .......... .......... .......... 60% 5.87M 9s
27000K .......... .......... .......... .......... .......... 60% 5.88M 9s
27050K .......... .......... .......... .......... .......... 60% 4.03M 9s
27100K .......... .......... .......... .......... .......... 60% 216K 9s
27150K .......... .......... .......... .......... .......... 60% 3.90M 9s
27200K .......... .......... .......... .......... .......... 60% 3.86M 9s
27250K .......... .......... .......... .......... .......... 60% 3.80M 9s
27300K .......... .......... .......... .......... .......... 60% 3.96M 9s
27350K .......... .......... .......... .......... .......... 61% 5.83M 9s
27400K .......... .......... .......... .......... .......... 61% 5.70M 9s
27450K .......... .......... .......... .......... .......... 61% 3.92M 9s
27500K .......... .......... .......... .......... .......... 61% 5.90M 9s
27550K .......... .......... .......... .......... .......... 61% 5.83M 9s
27600K .......... .......... .......... .......... .......... 61% 5.71M 9s
27650K .......... .......... .......... .......... .......... 61% 10.5M 9s
27700K .......... .......... .......... .......... .......... 61% 5.98M 9s
27750K .......... .......... .......... .......... .......... 61% 11.1M 9s
27800K .......... .......... .......... .......... .......... 62% 5.92M 9s
27850K .......... .......... .......... .......... .......... 62% 5.91M 9s
27900K .......... .......... .......... .......... .......... 62% 10.7M 9s
27950K .......... .......... .......... .......... .......... 62% 10.9M 8s
28000K .......... .......... .......... .......... .......... 62% 6.21M 8s
28050K .......... .......... .......... .......... .......... 62% 11.0M 8s
28100K .......... .......... .......... .......... .......... 62% 12.1M 8s
28150K .......... .......... .......... .......... .......... 62% 11.2M 8s
28200K .......... .......... .......... .......... .......... 62% 6.04M 8s
28250K .......... .......... .......... .......... .......... 63% 10.9M 8s
28300K .......... .......... .......... .......... .......... 63% 11.3M 8s
28350K .......... .......... .......... .......... .......... 63% 12.2M 8s
28400K .......... .......... .......... .......... .......... 63% 6.26M 8s
28450K .......... .......... .......... .......... .......... 63% 12.0M 8s
28500K .......... .......... .......... .......... .......... 63% 11.5M 8s
28550K .......... .......... .......... .......... .......... 63% 11.9M 8s
28600K .......... .......... .......... .......... .......... 63% 11.0M 8s
28650K .......... .......... .......... .......... .......... 63% 11.9M 8s
28700K .......... .......... .......... .......... .......... 64% 11.6M 8s
28750K .......... .......... .......... .......... .......... 64% 11.8M 8s
28800K .......... .......... .......... .......... .......... 64% 11.0M 8s
28850K .......... .......... .......... .......... .......... 64% 11.4M 8s
28900K .......... .......... .......... .......... .......... 64% 13.1M 8s
28950K .......... .......... .......... .......... .......... 64% 11.7M 8s
29000K .......... .......... .......... .......... .......... 64% 11.3M 8s
29050K .......... .......... .......... .......... .......... 64% 12.5M 8s
29100K .......... .......... .......... .......... .......... 64% 12.0M 8s
29150K .......... .......... .......... .......... .......... 65% 11.1M 8s
29200K .......... .......... .......... .......... .......... 65% 76.5M 8s
29250K .......... .......... .......... .......... .......... 65% 12.2M 8s
29300K .......... .......... .......... .......... .......... 65% 11.2M 8s
29350K .......... .......... .......... .......... .......... 65% 13.0M 7s
29400K .......... .......... .......... .......... .......... 65% 13.3M 7s
29450K .......... .......... .......... .......... .......... 65% 9.92M 7s
29500K .......... .......... .......... .......... .......... 65% 15.0M 7s
29550K .......... .......... .......... .......... .......... 65% 53.4M 7s
29600K .......... .......... .......... .......... .......... 66% 11.9M 7s
29650K .......... .......... .......... .......... .......... 66% 13.3M 7s
29700K .......... .......... .......... .......... .......... 66% 10.3M 7s
29750K .......... .......... .......... .......... .......... 66% 13.8M 7s
29800K .......... .......... .......... .......... .......... 66% 62.2M 7s
29850K .......... .......... .......... .......... .......... 66% 12.2M 7s
29900K .......... .......... .......... .......... .......... 66% 392K 7s
29950K .......... .......... .......... .......... .......... 66% 3.09M 7s
30000K .......... .......... .......... .......... .......... 66% 5.60M 7s
30050K .......... .......... .......... .......... .......... 67% 12.3M 7s
30100K .......... .......... .......... .......... .......... 67% 11.5M 7s
30150K .......... .......... .......... .......... .......... 67% 12.1M 7s
30200K .......... .......... .......... .......... .......... 67% 689K 7s
30250K .......... .......... .......... .......... .......... 67% 5.87M 7s
30300K .......... .......... .......... .......... .......... 67% 5.92M 7s
30350K .......... .......... .......... .......... .......... 67% 11.1M 7s
30400K .......... .......... .......... .......... .......... 67% 5.57M 7s
30450K .......... .......... .......... .......... .......... 67% 10.7M 7s
30500K .......... .......... .......... .......... .......... 68% 11.2M 7s
30550K .......... .......... .......... .......... .......... 68% 10.2M 7s
30600K .......... .......... .......... .......... .......... 68% 6.54M 7s
30650K .......... .......... .......... .......... .......... 68% 10.8M 7s
30700K .......... .......... .......... .......... .......... 68% 10.6M 7s
30750K .......... .......... .......... .......... .......... 68% 6.17M 7s
30800K .......... .......... .......... .......... .......... 68% 11.5M 7s
30850K .......... .......... .......... .......... .......... 68% 10.3M 7s
30900K .......... .......... .......... .......... .......... 68% 11.6M 7s
30950K .......... .......... .......... .......... .......... 69% 11.6M 7s
31000K .......... .......... .......... .......... .......... 69% 10.4M 6s
31050K .......... .......... .......... .......... .......... 69% 11.7M 6s
31100K .......... .......... .......... .......... .......... 69% 11.9M 6s
31150K .......... .......... .......... .......... .......... 69% 11.9M 6s
31200K .......... .......... .......... .......... .......... 69% 10.9M 6s
31250K .......... .......... .......... .......... .......... 69% 11.2M 6s
31300K .......... .......... .......... .......... .......... 69% 11.3M 6s
31350K .......... .......... .......... .......... .......... 69% 11.3M 6s
31400K .......... .......... .......... .......... .......... 70% 11.6M 6s
31450K .......... .......... .......... .......... .......... 70% 11.5M 6s
31500K .......... .......... .......... .......... .......... 70% 11.8M 6s
31550K .......... .......... .......... .......... .......... 70% 12.1M 6s
31600K .......... .......... .......... .......... .......... 70% 11.1M 6s
31650K .......... .......... .......... .......... .......... 70% 12.1M 6s
31700K .......... .......... .......... .......... .......... 70% 10.8M 6s
31750K .......... .......... .......... .......... .......... 70% 14.0M 6s
31800K .......... .......... .......... .......... .......... 70% 12.0M 6s
31850K .......... .......... .......... .......... .......... 71% 11.5M 6s
31900K .......... .......... .......... .......... .......... 71% 40.4M 6s
31950K .......... .......... .......... .......... .......... 71% 12.8M 6s
32000K .......... .......... .......... .......... .......... 71% 11.4M 6s
32050K .......... .......... .......... .......... .......... 71% 11.8M 6s
32100K .......... .......... .......... .......... .......... 71% 12.4M 6s
32150K .......... .......... .......... .......... .......... 71% 61.0M 6s
32200K .......... .......... .......... .......... .......... 71% 12.3M 6s
32250K .......... .......... .......... .......... .......... 71% 11.7M 6s
32300K .......... .......... .......... .......... .......... 72% 12.8M 6s
32350K .......... .......... .......... .......... .......... 72% 12.8M 6s
32400K .......... .......... .......... .......... .......... 72% 43.1M 6s
32450K .......... .......... .......... .......... .......... 72% 12.0M 6s
32500K .......... .......... .......... .......... .......... 72% 14.5M 6s
32550K .......... .......... .......... .......... .......... 72% 11.7M 6s
32600K .......... .......... .......... .......... .......... 72% 56.1M 5s
32650K .......... .......... .......... .......... .......... 72% 12.5M 5s
32700K .......... .......... .......... .......... .......... 72% 13.3M 5s
32750K .......... .......... .......... .......... .......... 73% 12.5M 5s
32800K .......... .......... .......... .......... .......... 73% 73.6M 5s
32850K .......... .......... .......... .......... .......... 73% 11.5M 5s
32900K .......... .......... .......... .......... .......... 73% 12.9M 5s
32950K .......... .......... .......... .......... .......... 73% 12.0M 5s
33000K .......... .......... .......... .......... .......... 73% 69.2M 5s
33050K .......... .......... .......... .......... .......... 73% 138K 5s
33100K .......... .......... .......... .......... .......... 73% 625K 5s
33150K .......... .......... .......... .......... .......... 73% 61.7M 5s
33200K .......... .......... .......... .......... .......... 74% 6.76M 5s
33250K .......... .......... .......... .......... .......... 74% 10.8M 5s
33300K .......... .......... .......... .......... .......... 74% 6.10M 5s
33350K .......... .......... .......... .......... .......... 74% 11.0M 5s
33400K .......... .......... .......... .......... .......... 74% 2.81M 5s
33450K .......... .......... .......... .......... .......... 74% 11.0M 5s
33500K .......... .......... .......... .......... .......... 74% 11.0M 5s
33550K .......... .......... .......... .......... .......... 74% 6.05M 5s
33600K .......... .......... .......... .......... .......... 74% 255K 5s
33650K .......... .......... .......... .......... .......... 75% 106K 5s
33700K .......... .......... .......... .......... .......... 75% 2.05M 5s
33750K .......... .......... .......... .......... .......... 75% 1.56M 5s
33800K .......... .......... .......... .......... .......... 75% 2.07M 5s
33850K .......... .......... .......... .......... .......... 75% 2.40M 5s
33900K .......... .......... .......... .......... .......... 75% 4.03M 5s
33950K .......... .......... .......... .......... .......... 75% 5.94M 5s
34000K .......... .......... .......... .......... .......... 75% 5.89M 5s
34050K .......... .......... .......... .......... .......... 75% 5.73M 5s
34100K .......... .......... .......... .......... .......... 76% 5.65M 5s
34150K .......... .......... .......... .......... .......... 76% 5.71M 5s
34200K .......... .......... .......... .......... .......... 76% 10.6M 5s
34250K .......... .......... .......... .......... .......... 76% 3.95M 5s
34300K .......... .......... .......... .......... .......... 76% 11.1M 5s
34350K .......... .......... .......... .......... .......... 76% 5.87M 5s
34400K .......... .......... .......... .......... .......... 76% 11.3M 5s
34450K .......... .......... .......... .......... .......... 76% 6.15M 5s
34500K .......... .......... .......... .......... .......... 76% 11.2M 5s
34550K .......... .......... .......... .......... .......... 77% 11.4M 5s
34600K .......... .......... .......... .......... .......... 77% 507K 5s
34650K .......... .......... .......... .......... .......... 77% 7.05M 5s
34700K .......... .......... .......... .......... .......... 77% 303K 5s
34750K .......... .......... .......... .......... .......... 77% 76.8K 5s
34800K .......... .......... .......... .......... .......... 77% 25.0K 5s
34850K .......... .......... .......... .......... .......... 77% 20.6K 6s
34900K .......... .......... .......... .......... .......... 77% 24.9K 7s
34950K .......... .......... .......... .......... .......... 77% 32.2K 7s
35000K .......... .......... .......... .......... .......... 78% 342K 7s
35050K .......... .......... .......... .......... .......... 78% 2.93M 7s
35100K .......... .......... .......... .......... .......... 78% 710K 7s
35150K .......... .......... .......... .......... .......... 78% 2.84M 7s
35200K .......... .......... .......... .......... .......... 78% 3.83M 7s
35250K .......... .......... .......... .......... .......... 78% 3.83M 7s
35300K .......... .......... .......... .......... .......... 78% 5.75M 7s
35350K .......... .......... .......... .......... .......... 78% 3.93M 7s
35400K .......... .......... .......... .......... .......... 78% 5.78M 7s
35450K .......... .......... .......... .......... .......... 79% 3.94M 7s
35500K .......... .......... .......... .......... .......... 79% 5.72M 7s
35550K .......... .......... .......... .......... .......... 79% 9.21M 7s
35600K .......... .......... .......... .......... .......... 79% 7.00M 7s
35650K .......... .......... .......... .......... .......... 79% 6.46M 7s
35700K .......... .......... .......... .......... .......... 79% 9.02M 6s
35750K .......... .......... .......... .......... .......... 79% 11.2M 6s
35800K .......... .......... .......... .......... .......... 79% 7.35M 6s
35850K .......... .......... .......... .......... .......... 79% 8.65M 6s
35900K .......... .......... .......... .......... .......... 80% 7.56M 6s
35950K .......... .......... .......... .......... .......... 80% 12.1M 6s
36000K .......... .......... .......... .......... .......... 80% 7.29M 6s
36050K .......... .......... .......... .......... .......... 80% 11.2M 6s
36100K .......... .......... .......... .......... .......... 80% 11.6M 6s
36150K .......... .......... .......... .......... .......... 80% 12.4M 6s
36200K .......... .......... .......... .......... .......... 80% 6.52M 6s
36250K .......... .......... .......... .......... .......... 80% 11.5M 6s
36300K .......... .......... .......... .......... .......... 80% 11.6M 6s
36350K .......... .......... .......... .......... .......... 81% 12.0M 6s
36400K .......... .......... .......... .......... .......... 81% 12.9M 6s
36450K .......... .......... .......... .......... .......... 81% 12.3M 6s
36500K .......... .......... .......... .......... .......... 81% 11.5M 6s
36550K .......... .......... .......... .......... .......... 81% 12.4M 6s
36600K .......... .......... .......... .......... .......... 81% 13.2M 6s
36650K .......... .......... .......... .......... .......... 81% 6.11M 6s
36700K .......... .......... .......... .......... .......... 81% 11.9M 6s
36750K .......... .......... .......... .......... .......... 81% 11.9M 6s
36800K .......... .......... .......... .......... .......... 82% 58.9M 6s
36850K .......... .......... .......... .......... .......... 82% 12.4M 6s
36900K .......... .......... .......... .......... .......... 82% 11.5M 5s
36950K .......... .......... .......... .......... .......... 82% 8.63M 5s
37000K .......... .......... .......... .......... .......... 82% 11.9M 5s
37050K .......... .......... .......... .......... .......... 82% 10.5M 5s
37100K .......... .......... .......... .......... .......... 82% 13.3M 5s
37150K .......... .......... .......... .......... .......... 82% 11.8M 5s
37200K .......... .......... .......... .......... .......... 82% 12.2M 5s
37250K .......... .......... .......... .......... .......... 83% 76.1M 5s
37300K .......... .......... .......... .......... .......... 83% 11.5M 5s
37350K .......... .......... .......... .......... .......... 83% 12.5M 5s
37400K .......... .......... .......... .......... .......... 83% 12.6M 5s
37450K .......... .......... .......... .......... .......... 83% 3.93M 5s
37500K .......... .......... .......... .......... .......... 83% 11.1M 5s
37550K .......... .......... .......... .......... .......... 83% 12.1M 5s
37600K .......... .......... .......... .......... .......... 83% 13.0M 5s
37650K .......... .......... .......... .......... .......... 83% 51.0M 5s
37700K .......... .......... .......... .......... .......... 84% 11.2M 5s
37750K .......... .......... .......... .......... .......... 84% 13.2M 5s
37800K .......... .......... .......... .......... .......... 84% 126K 5s
37850K .......... .......... .......... .......... .......... 84% 497K 5s
37900K .......... .......... .......... .......... .......... 84% 10.6M 5s
37950K .......... .......... .......... .......... .......... 84% 519K 5s
38000K .......... .......... .......... .......... .......... 84% 124K 5s
38050K .......... .......... .......... .......... .......... 84% 11.6M 5s
38100K .......... .......... .......... .......... .......... 84% 384K 5s
38150K .......... .......... .......... .......... .......... 85% 136K 5s
38200K .......... .......... .......... .......... .......... 85% 124K 5s
38250K .......... .......... .......... .......... .......... 85% 59.6K 5s
38300K .......... .......... .......... .......... .......... 85% 5.63M 5s
38350K .......... .......... .......... .......... .......... 85% 93.8K 5s
38400K .......... .......... .......... .......... .......... 85% 149K 5s
38450K .......... .......... .......... .......... .......... 85% 2.90M 5s
38500K .......... .......... .......... .......... .......... 85% 3.84M 5s
38550K .......... .......... .......... .......... .......... 85% 3.75M 5s
38600K .......... .......... .......... .......... .......... 86% 5.79M 5s
38650K .......... .......... .......... .......... .......... 86% 3.91M 5s
38700K .......... .......... .......... .......... .......... 86% 11.5M 5s
38750K .......... .......... .......... .......... .......... 86% 6.18M 5s
38800K .......... .......... .......... .......... .......... 86% 10.4M 5s
38850K .......... .......... .......... .......... .......... 86% 11.7M 5s
38900K .......... .......... .......... .......... .......... 86% 12.5M 4s
38950K .......... .......... .......... .......... .......... 86% 11.4M 4s
39000K .......... .......... .......... .......... .......... 86% 13.4M 4s
39050K .......... .......... .......... .......... .......... 87% 26.5M 4s
39100K .......... .......... .......... .......... .......... 87% 18.7M 4s
39150K .......... .......... .......... .......... .......... 87% 12.6M 4s
39200K .......... .......... .......... .......... .......... 87% 30.5M 4s
39250K .......... .......... .......... .......... .......... 87% 15.6M 4s
39300K .......... .......... .......... .......... .......... 87% 41.8M 4s
39350K .......... .......... .......... .......... .......... 87% 16.7M 4s
39400K .......... .......... .......... .......... .......... 87% 34.3M 4s
39450K .......... .......... .......... .......... .......... 87% 16.1M 4s
39500K .......... .......... .......... .......... .......... 88% 16.4M 4s
39550K .......... .......... .......... .......... .......... 88% 38.0M 4s
39600K .......... .......... .......... .......... .......... 88% 16.6M 4s
39650K .......... .......... .......... .......... .......... 88% 31.5M 4s
39700K .......... .......... .......... .......... .......... 88% 17.9M 4s
39750K .......... .......... .......... .......... .......... 88% 38.0M 4s
39800K .......... .......... .......... .......... .......... 88% 19.6M 4s
39850K .......... .......... .......... .......... .......... 88% 28.5M 4s
39900K .......... .......... .......... .......... .......... 88% 18.0M 4s
39950K .......... .......... .......... .......... .......... 89% 14.1M 4s
40000K .......... .......... .......... .......... .......... 89% 32.8M 4s
40050K .......... .......... .......... .......... .......... 89% 17.6M 4s
40100K .......... .......... .......... .......... .......... 89% 37.7M 3s
40150K .......... .......... .......... .......... .......... 89% 17.7M 3s
40200K .......... .......... .......... .......... .......... 89% 42.2M 3s
40250K .......... .......... .......... .......... .......... 89% 15.2M 3s
40300K .......... .......... .......... .......... .......... 89% 47.3M 3s
40350K .......... .......... .......... .......... .......... 89% 16.7M 3s
40400K .......... .......... .......... .......... .......... 90% 44.9M 3s
40450K .......... .......... .......... .......... .......... 90% 16.6M 3s
40500K .......... .......... .......... .......... .......... 90% 35.6M 3s
40550K .......... .......... .......... .......... .......... 90% 18.5M 3s
40600K .......... .......... .......... .......... .......... 90% 43.4M 3s
40650K .......... .......... .......... .......... .......... 90% 8.45M 3s
40700K .......... .......... .......... .......... .......... 90% 75.7M 3s
40750K .......... .......... .......... .......... .......... 90% 6.56M 3s
40800K .......... .......... .......... .......... .......... 90% 77.8M 3s
40850K .......... .......... .......... .......... .......... 91% 12.8M 3s
40900K .......... .......... .......... .......... .......... 91% 12.6M 3s
40950K .......... .......... .......... .......... .......... 91% 76.6M 3s
41000K .......... .......... .......... .......... .......... 91% 12.0M 3s
41050K .......... .......... .......... .......... .......... 91% 12.7M 3s
41100K .......... .......... .......... .......... .......... 91% 12.6M 3s
41150K .......... .......... .......... .......... .......... 91% 62.8M 3s
41200K .......... .......... .......... .......... .......... 91% 13.5M 3s
41250K .......... .......... .......... .......... .......... 91% 61.5M 3s
41300K .......... .......... .......... .......... .......... 92% 13.5M 3s
41350K .......... .......... .......... .......... .......... 92% 12.1M 2s
41400K .......... .......... .......... .......... .......... 92% 70.3M 2s
41450K .......... .......... .......... .......... .......... 92% 12.1M 2s
41500K .......... .......... .......... .......... .......... 92% 16.0M 2s
41550K .......... .......... .......... .......... .......... 92% 4.60M 2s
41600K .......... .......... .......... .......... .......... 92% 67.4M 2s
41650K .......... .......... .......... .......... .......... 92% 12.9M 2s
41700K .......... .......... .......... .......... .......... 92% 12.6M 2s
41750K .......... .......... .......... .......... .......... 93% 48.8M 2s
41800K .......... .......... .......... .......... .......... 93% 16.1M 2s
41850K .......... .......... .......... .......... .......... 93% 49.1M 2s
41900K .......... .......... .......... .......... .......... 93% 12.6M 2s
41950K .......... .......... .......... .......... .......... 93% 12.8M 2s
42000K .......... .......... .......... .......... .......... 93% 63.3M 2s
42050K .......... .......... .......... .......... .......... 93% 12.9M 2s
42100K .......... .......... .......... .......... .......... 93% 73.3M 2s
42150K .......... .......... .......... .......... .......... 93% 13.7M 2s
42200K .......... .......... .......... .......... .......... 94% 13.2M 2s
42250K .......... .......... .......... .......... .......... 94% 47.8M 2s
42300K .......... .......... .......... .......... .......... 94% 12.0M 2s
42350K .......... .......... .......... .......... .......... 94% 66.3M 2s
42400K .......... .......... .......... .......... .......... 94% 14.1M 2s
42450K .......... .......... .......... .......... .......... 94% 67.6M 2s
42500K .......... .......... .......... .......... .......... 94% 14.5M 2s
42550K .......... .......... .......... .......... .......... 94% 12.7M 2s
42600K .......... .......... .......... .......... .......... 94% 65.3M 2s
42650K .......... .......... .......... .......... .......... 95% 13.0M 2s
42700K .......... .......... .......... .......... .......... 95% 77.4M 1s
42750K .......... .......... .......... .......... .......... 95% 13.2M 1s
42800K .......... .......... .......... .......... .......... 95% 73.2M 1s
42850K .......... .......... .......... .......... .......... 95% 13.5M 1s
42900K .......... .......... .......... .......... .......... 95% 12.9M 1s
42950K .......... .......... .......... .......... .......... 95% 66.7M 1s
43000K .......... .......... .......... .......... .......... 95% 14.3M 1s
43050K .......... .......... .......... .......... .......... 95% 56.6M 1s
43100K .......... .......... .......... .......... .......... 96% 13.5M 1s
43150K .......... .......... .......... .......... .......... 96% 76.3M 1s
43200K .......... .......... .......... .......... .......... 96% 12.8M 1s
43250K .......... .......... .......... .......... .......... 96% 66.5M 1s
43300K .......... .......... .......... .......... .......... 96% 14.4M 1s
43350K .......... .......... .......... .......... .......... 96% 69.8M 1s
43400K .......... .......... .......... .......... .......... 96% 13.9M 1s
43450K .......... .......... .......... .......... .......... 96% 13.4M 1s
43500K .......... .......... .......... .......... .......... 96% 56.5M 1s
43550K .......... .......... .......... .......... .......... 97% 14.5M 1s
43600K .......... .......... .......... .......... .......... 97% 67.3M 1s
43650K .......... .......... .......... .......... .......... 97% 13.6M 1s
43700K .......... .......... .......... .......... .......... 97% 78.9M 1s
43750K .......... .......... .......... .......... .......... 97% 13.2M 1s
43800K .......... .......... .......... .......... .......... 97% 68.9M 1s
43850K .......... .......... .......... .......... .......... 97% 12.9M 1s
43900K .......... .......... .......... .......... .......... 97% 66.4M 1s
43950K .......... .......... .......... .......... .......... 97% 14.0M 1s
44000K .......... .......... .......... .......... .......... 98% 63.3M 1s
44050K .......... .......... .......... .......... .......... 98% 14.0M 1s
44100K .......... .......... .......... .......... .......... 98% 65.0M 1s
44150K .......... .......... .......... .......... .......... 98% 13.6M 0s
44200K .......... .......... .......... .......... .......... 98% 62.7M 0s
44250K .......... .......... .......... .......... .......... 98% 14.0M 0s
44300K .......... .......... .......... .......... .......... 98% 67.0M 0s
44350K .......... .......... .......... .......... .......... 98% 13.9M 0s
44400K .......... .......... .......... .......... .......... 98% 54.4M 0s
44450K .......... .......... .......... .......... .......... 99% 16.1M 0s
44500K .......... .......... .......... .......... .......... 99% 56.0M 0s
44550K .......... .......... .......... .......... .......... 99% 14.2M 0s
44600K .......... .......... .......... .......... .......... 99% 55.1M 0s
44650K .......... .......... .......... .......... .......... 99% 13.8M 0s
44700K .......... .......... .......... .......... .......... 99% 64.7M 0s
44750K .......... .......... .......... .......... .......... 99% 15.9M 0s
44800K .......... .......... .......... .......... .......... 99% 62.7M 0s
44850K .......... .......... .......... .......... .......... 99% 73.7M 0s
44900K ... 100% 7.37T=29s
Opening dataset from [oceanspy_particle_properties.nc].
2023-04-11 22:44:37 (1.49 MB/s) - ‘oceanspy_particle_properties.nc’ saved [45981653/45981653]
The OceanDataset created by extract_properties has 3 dimensions:
particle: Every particle is associated with an ID (integer number).time: All the available snapshots (in this case, from Feb. 29th to Mar. 30th with 6-hour resolution).time_midp: Mid-points of the time dimension. For example, this is the dimension associated with gradients computed along the time axis.
[7]:
print(od_lag.dataset)
<xarray.Dataset>
Dimensions: (time: 121, particle: 3954, time_midp: 120)
Coordinates:
* particle (particle) int64 0 1 2 3 4 5 6 ... 3948 3949 3950 3951 3952 3953
* time (time) datetime64[ns] 2008-02-29 ... 2008-03-30
* time_midp (time_midp) datetime64[ns] 2008-02-29T03:00:00 ... 2008-03-29T...
Data variables:
XC (time, particle) float64 ...
YC (time, particle) float64 ...
Z (time, particle) float64 ...
Zl (time, particle) float64 ...
Zp1 (time, particle) float64 ...
Zu (time, particle) float64 ...
XG (time, particle) float64 ...
YG (time, particle) float64 ...
Depth (time, particle) float64 ...
Temp (time, particle) float64 ...
S (time, particle) float64 ...
momVort3 (time, particle) float64 ...
Attributes:
OceanSpy_grid_coords: {'time': {'time': -0.5, 'time_midp': None}}
Compute
We can now use ospy.compute functions on the extracted particle properties. For example, here we compute potential density anomalies using temperature and salinity.
[8]:
od_lag = od_lag.compute.potential_density_anomaly()
print(od_lag.dataset["Sigma0"])
Computing potential density anomaly using the following parameters: {'eq_state': 'jmd95'}.
<xarray.DataArray 'Sigma0' (time: 121, particle: 3954)>
array([[28.06448144, 28.06448144, 28.06150693, ..., 27.60797582,
27.77204289, 27.82874751],
[28.06722492, 28.06416057, 28.06416057, ..., 27.62017588,
27.75539891, 27.82740934],
[28.06796263, 28.06796263, 28.06796263, ..., 27.60038398,
27.73746555, 27.82305911],
...,
[27.97167365, 27.88427005, 27.81440205, ..., 27.61308105,
27.61245962, 27.80288992],
[27.97205047, 27.8742948 , 27.81334513, ..., 27.62929107,
27.62997148, 27.80246744],
[27.97295942, 27.86978131, 27.81499219, ..., 27.63008246,
27.65160468, 27.80403993]])
Coordinates:
* particle (particle) int64 0 1 2 3 4 5 6 ... 3948 3949 3950 3951 3952 3953
* time (time) datetime64[ns] 2008-02-29 ... 2008-03-30
Attributes:
units: kg/m^3
long_name: potential density anomaly
OceanSpy_parameters: {'eq_state': 'jmd95'}
Here we compute gradients along the time dimension for each variable previously extracted:
[9]:
od_lag = od_lag.compute.gradient()
print(od_lag.dataset)
Computing gradient.
<xarray.Dataset>
Dimensions: (time: 121, particle: 3954, time_midp: 120)
Coordinates:
* particle (particle) int64 0 1 2 3 4 5 ... 3949 3950 3951 3952 3953
* time (time) datetime64[ns] 2008-02-29 ... 2008-03-30
* time_midp (time_midp) datetime64[ns] 2008-02-29T03:00:00 ... 2008-...
Data variables: (12/26)
XC (time, particle) float64 -24.62 -24.62 ... -29.45 -31.48
YC (time, particle) float64 65.49 65.49 65.49 ... 67.82 66.79
Z (time, particle) float64 -23.5 -23.5 ... -276.5 -351.5
Zl (time, particle) float64 -27.0 -27.0 ... -269.0 -344.0
Zp1 (time, particle) float64 -27.0 -27.0 ... -269.0 -344.0
Zu (time, particle) float64 -27.0 -27.0 ... -269.0 -344.0
... ...
dYG_dtime (time_midp, particle) float64 -8.841e-07 0.0 ... 0.0 0.0
dDepth_dtime (time_midp, particle) float64 4.819e-05 ... -0.000665
dTemp_dtime (time_midp, particle) float64 -3.942e-07 ... -1.581e-07
dS_dtime (time_midp, particle) float64 1.183e-07 ... 7.341e-08
dmomVort3_dtime (time_midp, particle) float64 4.739e-10 ... -7.566e-10
dSigma0_dtime (time_midp, particle) float64 1.27e-07 ... 7.28e-08
Attributes:
OceanSpy_grid_coords: {'time': {'time': -0.5, 'time_midp': None}}
Mask and Split
Now we focus on particles carried by the dense overflow water, which is defined as water with \(\sigma_{0}\geq\) 27.8 kg m\(^{-3}\).
First, we identify all the particles seeded in the overflow. Then, we split these particles into two sets (each one an OceanDataset) using the following criteria:
od_dense: \(\sigma_0\) along the path is always greater than 27.8 kg m\(^{-3}\).od_mixed: \(\sigma_0\) at the last snapshot is less than 27.8 kg m\(^{-3}\), and at least 50% of the snapshots have \(\sigma_0<\) 27.8 kg m\(^{-3}\).
[10]:
# Overflow mask
DSO_threshold = 27.8 # kg/m^3
mask_DSO = xr.where(od_lag.dataset["Sigma0"] >= DSO_threshold, 1, 0)
# Mask particles always in the overflow
mask_dense = xr.where(mask_DSO.sum("time") == len(od_lag.dataset["time"]), 1, 0)
# Mask mixed particles
mask_mixed = xr.where(
np.logical_and(
np.logical_and(mask_DSO.isel(time=0) == 1, mask_DSO.isel(time=-1) == 0),
mask_DSO.sum("time") <= len(od_lag.dataset["time"]) / 2,
),
1,
0,
)
# Create OceanDataset
od_dense = ospy.OceanDataset(od_lag.dataset.where(mask_dense, drop=True))
od_mixed = ospy.OceanDataset(od_lag.dataset.where(mask_mixed, drop=True))
Here we plot the trajectories on top of projected maps:
[11]:
ods = [od_dense, od_mixed]
colors = ["fuchsia", "lime"]
titles = [
r"Particles always denser than 27.8 kg m$^{-3}$.",
r"Particles ONLY initially denser than 27.8 kg m$^{-3}$",
]
for _, (od_i, col, tit) in enumerate(zip(ods, colors, titles)):
# Plot Depth
fig = plt.figure(figsize=(10, 5))
ax = od_eul.plot.horizontal_section(varName="masked_Depth", cmap="bone_r")
land_col = (253 / 255, 180 / 255, 108 / 255)
ax.patch.set_facecolor(land_col)
ax.set_extent([-40, -19, 63, 68])
# Plot trajectories
ax.plot(
od_i.dataset["XC"],
od_i.dataset["YC"],
color=col,
alpha=0.2,
linewidth=1,
transform=PlateCarree(),
)
# Plot initial positions
ax.plot(
od_i.dataset["XC"].isel(time=0),
od_i.dataset["YC"].isel(time=0),
"k.",
markersize=5,
transform=PlateCarree(),
)
# Title
_ = ax.set_title(tit)
plt.show()
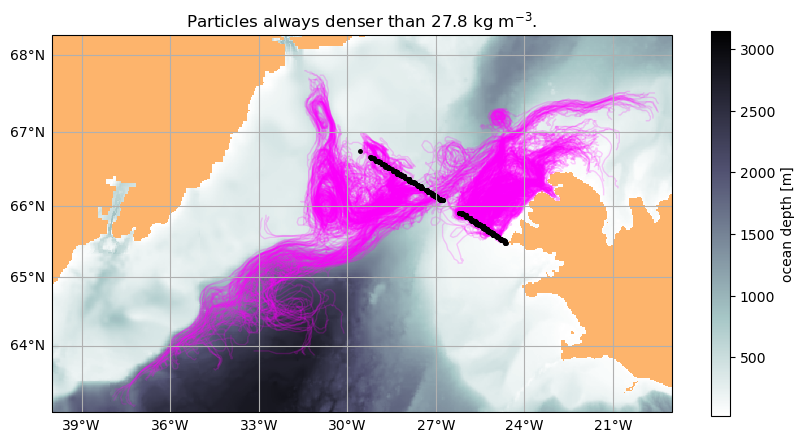
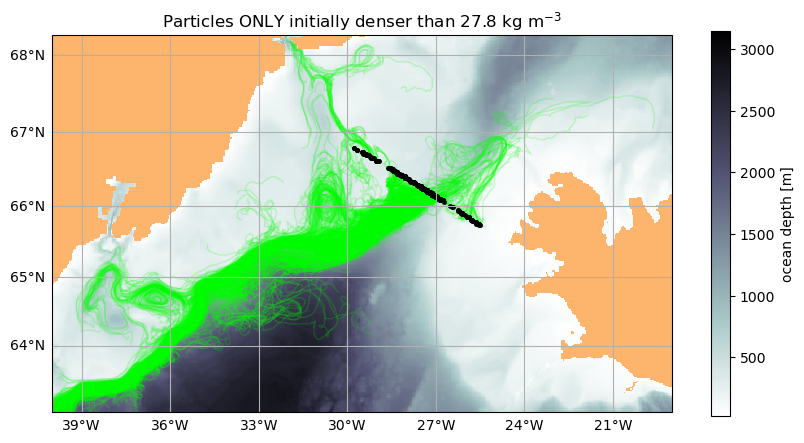
TS-diagrams
We can use TS-diagrams to investigate the hydrographic properties of the od_dense and od_mixed sets. For example, here we plot each oceandataset color-coded by particle depths separately (subplots a and b), then we plot them together on the same isopycnal contours (subplot c).
[12]:
fig = plt.figure(figsize=(10, 10))
for i, (od_i, col, tit) in enumerate(zip(ods, colors, titles)):
plt.subplot(int("22" + str(i + 1)))
ax = od_i.plot.TS_diagram(
colorName="Z", alpha=0.5, cmap_kwargs={"vmin": -2000, "vmax": 0}
)
ax.set_title(chr(ord("a") + i) + ") " + tit)
plt.subplot(212)
Tlim = [
min([od_i.dataset["Temp"].min().values for od_i in ods]),
max([od_i.dataset["Temp"].max().values for od_i in ods]),
]
Slim = [
min([od_i.dataset["S"].min().values for od_i in ods]),
max([od_i.dataset["S"].max().values for od_i in ods]),
]
for i, (od_i, col, tit) in enumerate(zip(ods, colors, titles)):
contour_kwargs = {}
if i == 0:
contour_kwargs["levels"] = []
ax = od_i.plot.TS_diagram(
color=col,
alpha=0.1,
Tlim=Tlim,
Slim=Slim,
contour_kwargs=contour_kwargs,
)
tit = "\n".join(
["c) "] + [f"{col}: {tit}" for _, (col, tit) in enumerate(zip(colors, titles))]
)
ax.set_title(tit)
plt.tight_layout()
Isopycnals: Computing potential density anomaly using the following parameters: {'eq_state': 'jmd95'}.
Isopycnals: Computing potential density anomaly using the following parameters: {'eq_state': 'jmd95'}.
Isopycnals: Computing potential density anomaly using the following parameters: {'eq_state': 'jmd95'}.
Isopycnals: Computing potential density anomaly using the following parameters: {'eq_state': 'jmd95'}.
/home/idies/mambaforge/envs/Oceanography/lib/python3.9/site-packages/oceanspy/plot.py:306: UserWarning: No contour levels were found within the data range.
CS = ax.contour(s.values, t.values, dens.values, **contour_kwargs)
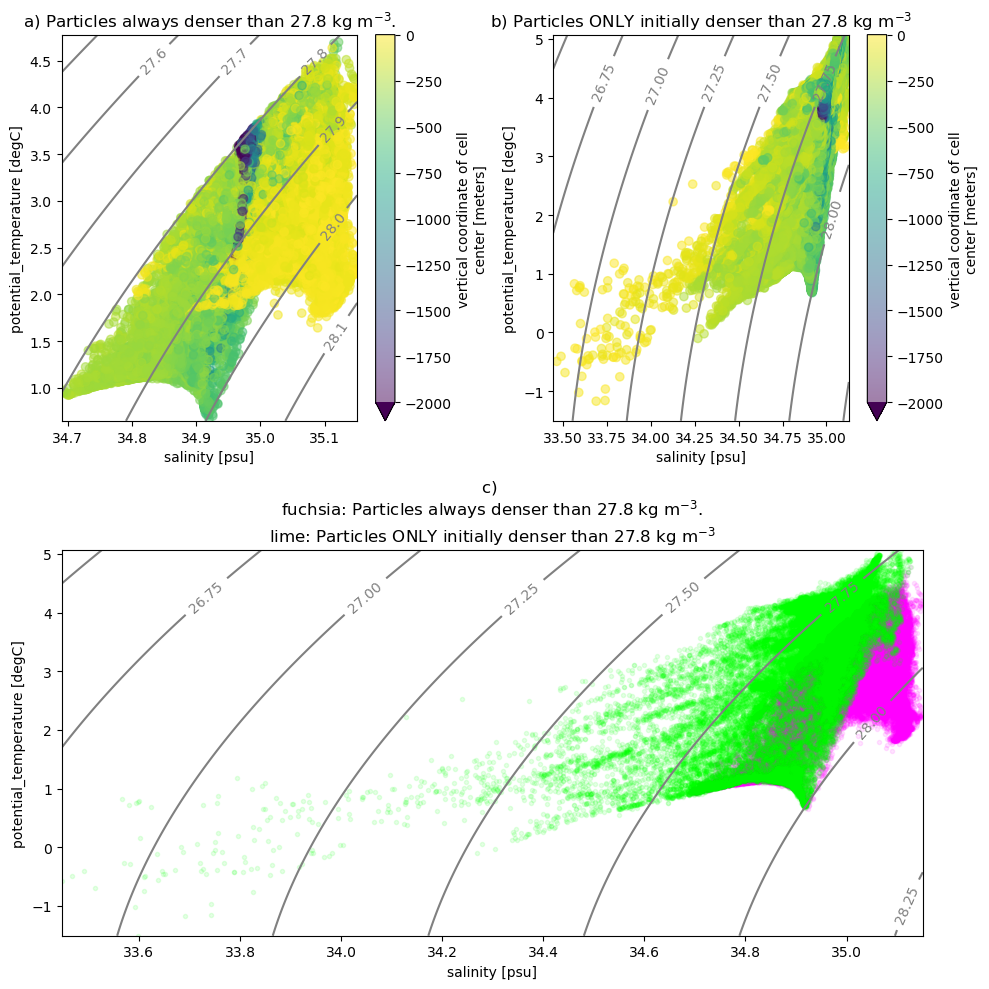
Time series
Now, we select 5 particles that were seeded in the overflow and became less dense along the path using the od_mixed set:
[13]:
nparts = 5
step = np.ceil(len(od_mixed.dataset["particle"]) / 5)
od_5parts = ospy.OceanDataset(
od_mixed.dataset.isel(particle=slice(None, None, int(step)))
)
Here is a plot of their trajectories:
[14]:
ods = [od_dense, od_mixed]
colors = ["fuchsia", "lime"]
titles = [
r"Particles always denser than 27.8 kg m$^{-3}$.",
r"Particles ONLY initially denser than 27.8 kg m$^{-3}$",
]
# Plot Depth
fig = plt.figure(figsize=(10, 5))
ax = od_eul.plot.horizontal_section(varName="masked_Depth", cmap="bone_r")
land_col = (253 / 255, 180 / 255, 108 / 255)
ax.patch.set_facecolor(land_col)
ax.set_extent([-40, -19, 63, 68])
# Plot trajectories
traj = ax.plot(
od_5parts.dataset["XC"],
od_5parts.dataset["YC"],
linewidth=3,
transform=PlateCarree(),
)
# Plot initial positions
i_pos = ax.plot(
od_5parts.dataset["XC"].isel(time=0),
od_5parts.dataset["YC"].isel(time=0),
"k.",
markersize=10,
transform=PlateCarree(),
)
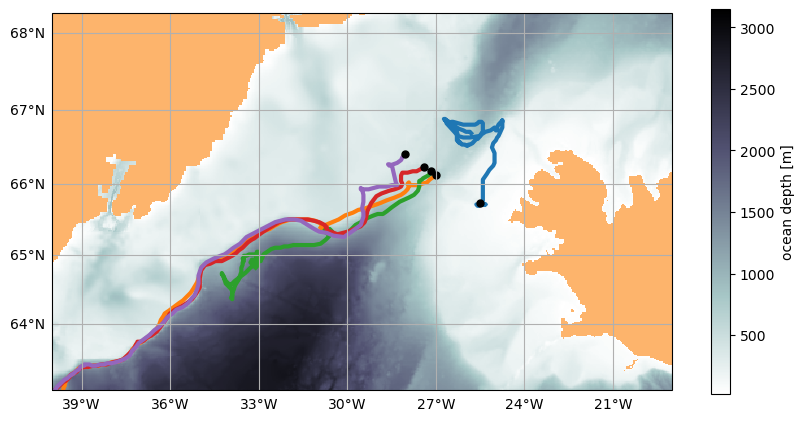
Now we plot the change in potential density anomaly as a function of time of these five particles (1st plot below), and the potential density anomaly gradient (2nd plot). The rapid decrease in particle density corresponds to intense mixing and entrainment of lighter ambient water as the overflow descends into the Irminger Sea. Notice one particle on the Iceland shelf (in blue) is not part of the overflow and moves north then mixes.
[15]:
varName = "Sigma0"
fig, axes = plt.subplots(2, 1, figsize=(10, 10), sharex=True)
for i, ax in enumerate(axes.flatten()):
if i == 1:
varName = f"d{varName}_dtime"
ax = od_5parts.plot.time_series(varName=varName, ax=ax, linewidth=2)
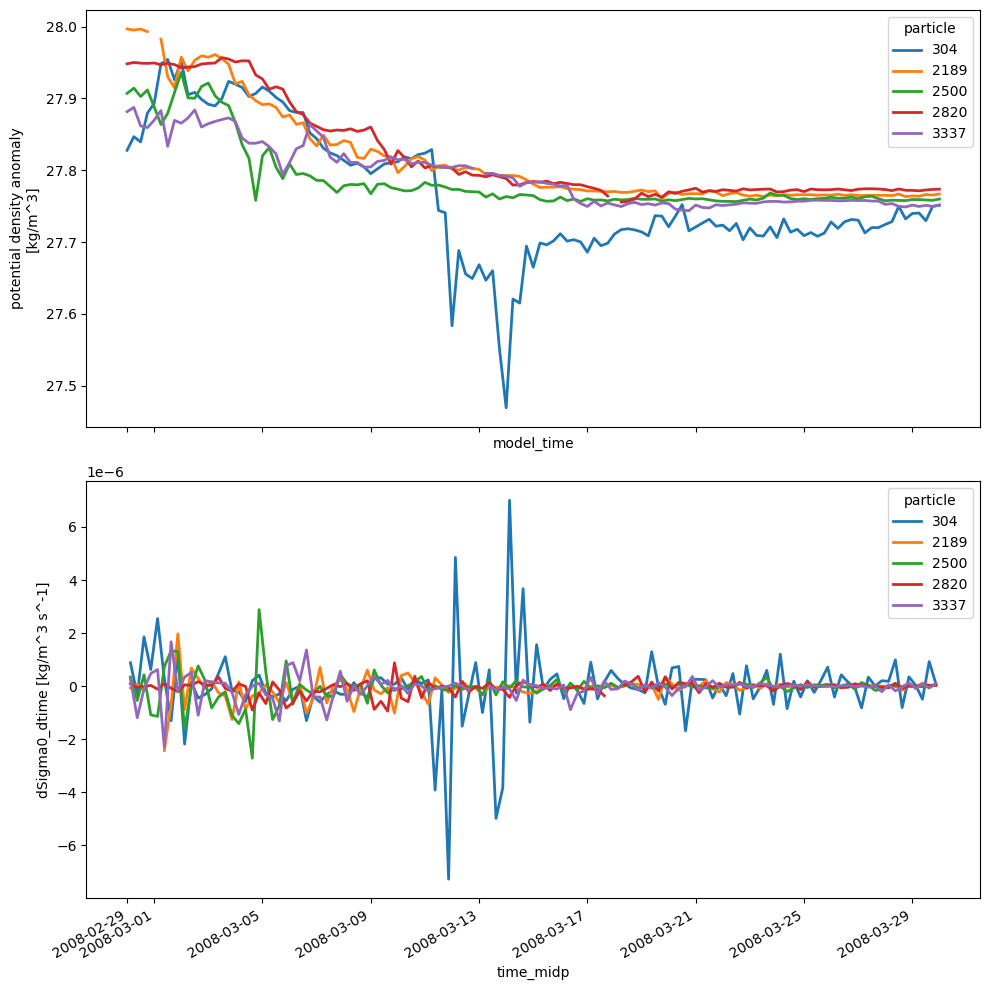
The plots above can be easily customized for any of the extracted variables. For example, change varName = 'Sigma0' in the cell above with any of the variables printed below.
[16]:
# Create table
table = {
var: od_5parts.dataset[var].attrs.pop(
"long_name", od_5parts.dataset[var].attrs.pop("description", None)
)
for var in od_5parts.dataset.variables
if "time" not in var and "particle" not in var
}
print("\n{:>15}: {}\n".format("varName", "label"))
for name, desc in sorted(table.items()):
print("{:>15}: {}".format(name, desc))
varName: label
Depth: ocean depth
S: salinity
Sigma0: potential density anomaly
Temp: potential_temperature
XC: longitude
XG: longitude
YC: latitude
YG: latitude
Z: vertical coordinate of cell center
Zl: vertical coordinate of upper cell interface
Zp1: vertical coordinate of cell interface
Zu: vertical coordinate of lower cell interface
momVort3: 3rd component (vertical) of Vorticity